Fetal weight estimation by ultrasound: development of Indian population-based models
Article information
Abstract
Purpose
Existing ultrasound-based fetal weight estimation models have been shown to have high errors when used in the Indian population. Therefore, the primary objective of this study was to develop Indian population-based models for fetal weight estimation, and the secondary objective was to compare their performance against established models.
Methods
Retrospectively collected data from 173 cases were used in this study. The inclusion criteria were a live singleton pregnancy and an interval from the ultrasound scan to delivery of ≤7 days. Multiple stepwise regression (MSR) and lasso regression methods were used to derive fetal weight estimation models using a randomly selected training group (n=137) with cross-products of abdominal circumference (AC), biparietal diameter (BPD), head circumference (HC), and femur length (FL) as independent variables. In the validation group (n=36), the bootstrap method was used to compare the performance of the new models against 12 existing models.
Results
The equations for the best-fit models obtained using the MSR and lasso methods were as follows: log10(EFW)=2.7843700+0.0004197(HC×AC)+0.0008545(AC×FL) and log10(EFW)=2.38 70211110+0.0074323216(HC)+0.0186555940(AC)+0.0013463735(BPD×FL)+0.0004519715 (HC×FL), respectively. In the training group, both models had very low systematic errors of 0.01% (±7.74%) and -0.03% (±7.70%), respectively. In the validation group, the performance of these models was found to be significantly better than that of the existing models.
Conclusion
The models presented in this study were found to be superior to existing models of ultrasound-based fetal weight estimation in the Indian population. We recommend a thorough evaluation of these models in independent studies.
Introduction
The accurate estimation of intrapartum fetal weight is very important for clinical management, as it is very closely linked with the survival and well-being of a fetus. Among the available intrapartum fetal weight estimation methods, ultrasound-based estimation is the most readily available and widely practiced technique. This has led to the development of a number of ultrasound-based models for fetal weight estimation. However, no single ultrasound-based model is found to be equally applicable to all populations [1], which could be due to differences across populations in genetic [2], anthropometric [3,4], nutritional, and socio-economic [5] factors that are known to impact fetal weight. Furthermore, studies have also pointed out that the use of racial/ethnic-specific standards improves the precision of fetal growth monitoring [6].
As such, a model developed on a particular population is most likely to work better for that population than for other populations. Consequently, it has been observed that existing models derived from non-Indian populations have high error when used in the Indian population [7]. Erroneous assessments made using such models could lead to missed or unnecessary interventions with deleterious effects on the fetus and mother. Differences in underlying population characteristics could contribute to these errors [7]. Using Indian population-based models would likely be beneficial in this scenario. Unfortunately, no Indian population-based models are yet available for this purpose; therefore, the primary aim of this study was to derive Indian-population based models for ultrasound-based fetal weight estimation. The secondary aim was to compare the performance of the newly generated models with that of the existing models.
Materials and Methods
A de-identified database (from 2013) of pregnant women obtained from a tertiary care hospital in Bengaluru (Bangalore), India was used for this retrospective study. The records were scrutinized for inclusion and exclusion criteria. The inclusion criteria were a live-birth singleton pregnancy and an interval between the last ultrasound scan and delivery of less than or equal to 7 days. All the ultrasound scans were performed by experienced radiologists using standard protocols. The weight of each newborn baby was measured immediately after birth. The study population included preterm, term, and postterm fetuses, as well small for gestational age and large for gestational age babies. Cases with a suspected fetal malformation or anomaly were excluded to avoid bias in the weight estimation. Similarly, cases of postpartum maternal or neonatal death were also excluded. Cases with complications other than the exclusion criteria were included in the study. The retrospective data used for the study were obtained in accordance with local regulations after receiving written approval from the institutional review board; the need for patient consent was waived by the review board.
The study population (n=173) was randomly split into two subgroups: a training set (80% of the study population, n=137) and a validation set (n=36). The training set was used to derive new models for fetal weight estimation. The validation set was used to test the performance of the generated models, and to compare it against the performance of the existing fetal weight estimation models.
Methodology for Deriving the New Models
To derive the new models, the actual birth weight (ABW) of a newborn in grams was considered to be the dependent parameter, while four routinely used ultrasound-based fetal biometry parameters-abdominal circumference (AC), biparietal diameter (BPD), head circumference (HC), and femur length (FL) in centimeterswere used as independent variables. As fetal weight gain during intrauterine life is exponential in nature, it has been observed that ultrasound-based independent variables correlate most closely with log10-transformed values of birth weight [8]. Therefore, log10 of ABW was used as the dependent parameter for model derivation. To generate new features, up to cubic-term cross-products of the four independent variables were used. The resultant 142 feature combinations were then used to derive the new models. All feature combinations considered in earlier studies [7] were included in this feature set.
Most of the previous researchers used multiple stepwise regression (MSR) analysis with a certain predefined criterion such as the F-test, adjusted R2 , or Mallow’s Cp to select the model with the most appropriate subset of features for fetal weight estimation. Although computationally less demanding, this method has shown to be less generalizable for new data sets as it often leads to locally optimal solutions [9]; moreover, this method becomes impractical with an increase in the number of independent features [9]. To overcome this problem, several new methods, such as least absolute shrinkage and selection operator (lasso) regression, have been introduced in the machine learning field.
As in MSR, the lasso method starts with a full set of features, but with every run it reduces the contribution of less important features. When the contribution of a feature drops below a certain threshold, the value of its coefficient is set to 0, effectively leading to the removal of that feature from the model. This process allows the selection of strongly correlated features, ultimately culminating in a simple model with fewer but more interpretable features. This latter capability makes the lasso method appropriate for feature selection in models with a large number of independent variables. During model derivation, the lasso method also regularizes features by shrinking their coefficients. Regularization helps to avoid the problem of overfitting data and improves the overall predictive accuracy and generalizability of a model [10].
For this study, both MSR analysis and the lasso method were used to derive new models. This was done in order to compare the performance of these two methods. In the MSR method, the Akaike information criterion was used for forward selection of a candidate model that minimized information loss. In the lasso method, 10-fold cross-validation (CV) of the training data was used to derive models; the model with the least CV error was selected for final testing. For model selection, preference was given to models that were relatively simple but had well-generalizable performance on the test data. The models were generated using R (ver. 3.3.2) [11,12].
Selection of the Existing Models for Comparison
It is impractical to compare the performance of models derived from different populations with each other, but since these models are routinely used on the Indian population, the performance of the newly derived models was compared with the performance of existing models. For this comparison, only models that have been found to have a systematic error of ±10% in the Indian population in earlier studies [7] were selected. The validation data set was used for this comparison. All 12 selected models (Table 1) [13-20] and the new models were implemented in MATLAB (MATLAB 9. 0.0.341360, The MathWorks Inc., Natick, MA, USA). As the models of Warsof (AC-BPD) and Ott (AC-HC-FL) estimated fetal weight (EFW) in kilograms, the values were converted to grams before the analysis.
Statistical Analysis
For an EFW given by a model, the percentage error was calculated using the following equation:
The performance of the models was compared in terms of: (1) the mean percentage error (MPE) and its standard deviation, (2) the mean absolute percentage error (APE) and its standard deviation, (3) the coefficient of determination (R2 ) and the Pearson correlation coefficient, and (4) analysis of the proportions of EFW within ±10% of the ABW. The Bland-Altman plot method was used to assess the limits of agreement between the ABW and EFW given by the new models.
The MPE is a measure of the magnitude of systemic error in fetal weight estimation in a model using ABW as the ground truth; therefore, it was used as the primary parameter for comparison. Random error (standard deviation of the systematic error) indicates the impact of various acquisition-related factors, including equipment calibration, image quality, variations in measurement, operator experience and training on overall error in weight estimation [1]. The 1-sample Student t test was used to determine whether the MPE (systematic error) of a derived model was significantly different from 0. The new models were compared with each other using the paired 2-sample Student t test. One-way analysis of variance (ANOVA) test was used to compare the percentage errors between a new model and the existing models; for pairwise comparisons, the Tukey honest significant difference (HSD) test was used. For all comparisons, a P-value of <0.05 was considered to indicate statistical significance. The normality assumption was tested for all parameters before the application of statistical tests.
Due to the small number of samples in the validation set, the bootstrap technique was used to assess the generalizability of the newly derived models and their performance in comparison with existing models. The bootstrap technique is a standard resampling technique in statistical analysis for deriving a large number of datasets by random sampling from the original dataset with replacement. All performance-related statistical parameters are measured in each bootstrap-derived dataset. An analysis of the mean and standard error of the statistical parameters from these derived datasets is then used to estimate the overall accuracy of the statistical parameters. As the derived datasets have distributions similar to the original data, it becomes possible to make inferences about a population from a sample dataset [22]. In this study, 10,000 bootstrap sample datasets were used to compare the performance of the new models and with that of the existing models. All statistical analyses were performed in R and MATLAB.
Results
In total, 173 cases met the inclusion and exclusion criteria. Nulliparous women constituted 48.5% of the study population. The mean gestational age of the study population was 38.5 weeks (range, 34 to 43.3 weeks), with 25 preterm (14.4%) and six postterm births. The mean body mass index of the study population was 28.07 kg/m2 (±0.63 kg/m2 ). The mean birth weight of the study population was 2,732.20 g (±369.99 g) with a range of 1,400 to 3,700 g; low-birth-weight babies (ABW ≤2,500 g) constituted 27.8% (n=48) of the study population. The average duration between the ultrasound scan and delivery was 2.7 days; 62.8% of cases had an ultrasound scan done within 3 days before delivery. The relevant demographic characteristics of the training and validation groups are summarized in Table 2. The two groups were found to have comparable demographic characteristics by the independent-samples Student t test.
Performance of the New Models
Compared to earlier studies in which a limited number of feature combinations was studied, in this study we derived models using 142 combinations of features. The best-fit models obtained by MSR (model 1) and the lasso method (model 2) from the training group are shown in Table 3. Model 1 had cross-products of AC with HC and FL as features, whereas model 2 included different cross-products of four biometry parameters. Although a large number of feature combinations was used, it was found that the best-fit models were composed of simple cross-products of fundamental biometry parameters without any high-order derivations; this indicates that the basic features were closely related to fetal weight.
Model 1 had systematic error of 0.01% (±7.74%) on the training group, with a mean difference of -13.95 g (±210.29 g). The adjusted R2 for model 1 was 0.656, and using other combinations or adding more terms yielded no further significant improvements in the model. Model 1 had 81% of its estimations within ±10% of ABW. For the lasso method (model 2), the model with the least CV error was selected; this model had a systematic error of -0.03% (±7.70%) in the training group with a mean difference of -16.25 g (±209.62 g) and an adjusted R2 equal to 0.633. Model 2 had 82% of its estimations within ±10% of ABW. The systematic error of these two models was found not to be significantly different from 0 by the 1-sample Student t test. No statistically significant difference was observed in the performance of the two new models using the paired 2-sample Student t test. The accuracy of these two models on training data is summarized in Table 4. The Bland-Altman plots for the limits of agreement with 95% confidence intervals for model 1 and model 2 are presented in Fig. 1.

Bland-Altman plot of model 1 (A) and model 2 (B) on the training group.
ABW, actual birth weight; EFW, estimated fetal weight; SD, standard deviation.
The systematic error of the two new models on the training data was found to be lower than the error values reported by other studies for their index populations [14,15,19]. Both new models had random error of more than 7%; although random error values of less than 7% have been rarely reported, retrospective data collection could have contributed to the slightly higher random error in our study, even though this error was found to be comparable with the random error reported for other models [1].
Comparative Analysis of the Models
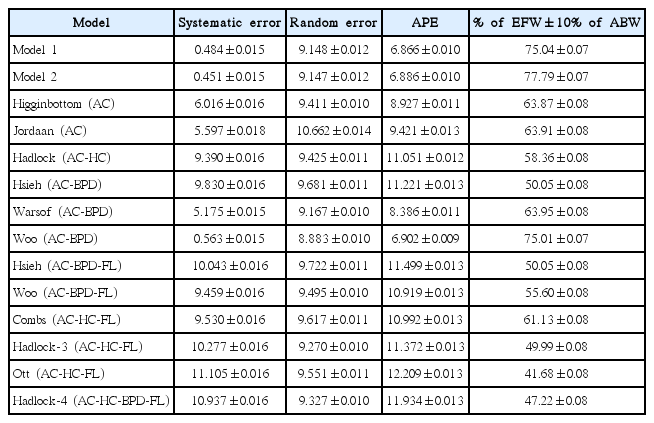
Based on the selection criteria described above, 12 existing models were selected for comparison with the two new models. For comparison, 10,000 bootstrap sample datasets derived from the validation group were used. We observed wide variation in the systematic error of the models, with a range from 0.45% to 11.01% (Table 5). Overall, the lowest systematic error (0.45%) was observed for model 2, closely followed by model 1. The difference in systematic error between the two new models and the existing models was found to be statistically significant by 1-way ANOVA. Subsequent pairwise comparison using the Tukey HSD test revealed that both new models had statistically significant less systematic error than all the existing models. Random error showed less variation than systematic error, with the Woo (AC-BPD) model having the lowest random error (8.88%), followed by model 2 and model 1. In regard to APE, model 1 had statistically significantly less error (6.87%) than all the other models, and model 2 also had significantly lower APE than all the other models except the Woo (AC-BPD) model. The best performance in terms of the highest number of predictions within ±10% of ABW was observed with model 2 (77.79%). The highest correlation coefficient (0.841) was observed with model 2 and the Combs (AC-HC-FL) model.
The overall performance of both new models was found to be significantly better than that of all the existing models, with lone exception of the Woo (AC-BPD) model. Both new models showed better performance than the Woo (AC-BPD) model for most performance measures as well, but the difference was not statistically significant for all measures.
Discussion
As a number of factors, including population characteristics, are known to impact fetal growth, it is very important to use appropriate models for fetal weight estimation. Nonetheless, no indigenous fetal weight estimation model has yet been developed for the Indian population. In the absence of such models, Indian practitioners must rely on other population-based models for fetal weight estimation. Unfortunately, such models are known to have high errors in the Indian population [7], thus putting both doctors and patients at a disadvantage. Therefore, the primary aim of this study was to derive Indian population-based models for fetal weight estimation.
In this study, models were derived based on two different methodologies: MSR and lasso regression. We observed that overall performance of these two new models was superior to the performance of the existing models, with low systematic error, random error, and APE; the new models also had high correlation coefficients and higher number of predictions within ±10% of ABW. The bootstrap technique (10,000 resamples) was used to empirically demonstrate that the performance of proposed models was better than that of the existing models, and would remain valid when used on samples other than the validation group [13]. Although no statistically significant difference was observed between the new models, the lasso regression-based model 2 was considered to be the more appropriate model for fetal weight estimation due to its low systematic error, relatively low random error, and higher number of predictions within ±10% of ABW. Among the existing models, we found the geographically closer, Hong Kong population-based Woo (AC-BPD) model to be the most appropriate model for our study population.
A number of studies have highlighted differences in fetal growth patterns between Indian and other populations. Those studies have observed that Indian fetuses have lower birth weight and are smaller in all body measurements [3,23]. This could be the reason for the observed weight overestimation by Western models when used for Indian babies [7]. Genetic factors are also known to influence fetal growth; researchers have observed that even second-generation immigrant mothers of Indian origin are likely to have babies with lower birth weight [2,24]. Considering these factors, our new models likely performed better because they were derived from an Indian population and the underlying population characteristics were better incorporated. However, it is important to note that when models are tested on the same population from which they are derived, there will be an inherent bias in favor of those models. Therefore, it is strongly recommended that the performance of these models should be validated in large independent studies.
Maternal factors that impact birth weight, such as diabetes mellitus, smoking, pregnancy-induced hypertension, and fetal factors such as gender are likely to affect a model’s performance. However, as we wanted to derive a general-purpose model, the entire birth weight range, with all maternal complications, was included in this study. Such general-purpose models offer a flexibility of having a single model across the whole range of weights and gestational ages rather than having multiple models, which may obscure the magnitude of altered fetal growth [25]. However, such models are known to have issues of weight overestimation in small fetuses and weight underestimation in large fetuses; as this issue has been consistently observed in all existing models [1,21], it needs due diligence from practitioners.
Considering the limitations of conventional ultrasound-based models in accurate fetal weight estimation, researchers have proposed including other parameters, such as mid-thigh soft tissue thickness [26] or maternal characteristics [27], in models. Studies have also proposed using volumetric methods based on 3-dimensional ultrasound or magnetic resonance imaging for fetal weight estimation [1]. Such models also need to be thoroughly validated for the Indian population before their application, as these models are also likely to be impacted by underlying population differences.
The retrospective design and a small sample size from a single center are two important limitations of our study. Although a smaller sample size of 137 cases was used for this study, it can be still considered comparable with the median sample number of previous studies. Another limiting factor is that we did not study the impact of other factors, such as maternal ethnicity, socioeconomic status, or geographic factors, which could have affected fetal weight. This makes it difficult to generalize the findings of this study for the entire country due to the prevailing geographical and ethnic diversity of India. Nevertheless, differences within India have been observed to be lesser in magnitude than differences between Indian and other populations [4].
To the best of our knowledge, this is the first time that an advanced method such as lasso regression has been used for a model derivation. The lasso method optimizes feature selection, thereby providing the model with fewer and more interpretable features. This makes it possible to explore a large number of feature combinations during model development. Moreover, this method also regularizes features by reducing their magnitude, which helps to avoid the problem of overfitting the data. Models resulting from this method have been shown to be more generalizable on newer datasets, with improved prediction capabilities [10] beyond what was hitherto possible with conventional methods such as MSR.
The main strength of our study lies in being the first study to present Indian population-based models for ultrasound-based fetal weight estimation. We observed that the overall performance of these models was superior to that of the existing models. These models are likely to be helpful to Indian clinicians by enabling better fetal weight estimation, which is expected to facilitate informed and timely decision-making. This is also the first study in which a state-of-the-art machine learning method such as lasso regression was used for model derivation. Given the advantages of this technique, we believe that this method could be helpful in developing more appropriate models for fetal weight estimation in the future. Considering the importance of fetal weight in clinical practice, it is further recommended that the models presented in this study should be validated with well-designed studies conducted throughout India.
Notes
No potential conflict of interest relevant to this article was reported.